NEMOTO Takahiro
Professor /PI
PRIMe, The University of Osaka
- Related Website:
- research gate X(Twitter) Personal Website
Our researchers
PRIMe researchers from diverse fields of study, nationalities, and backgrounds come together and collaborate “under-one-roof” to conduct interdisciplinary and integrative research.

NEMOTO Takahiro
Professor /PI
PRIMe, The University of Osaka
Feedback between Organoid Experiments and Mathematical Models
A key to a digital twin is close feedback between mathematical models and experiments. Through careful discussions with experimentalists, our team will develop algorithms that will serve as the building blocks for creating the human metaverse. One of our current goals is to develop a fine-tuned Polygenic Risk Score (PRS) using human organoid experimental data.
Research Outline
Previous work
Algorithm development and data analysis: In data analysis, it is important to discuss closely with the people who actually collect the data (through experiments, fieldwork, etc.) to develop appropriate algorithms for their problems. We have worked in close collaboration in various interdisciplinary areas such as fluid mechanics 1, biophysics 2, epidemiology 3, and bioinformatics 4. For example, French Polynesia is a beautiful tropical country in the Pacific Ocean, but it is plagued by mosquito-borne diseases such as dengue fever (Figure A). Dengue fever is known to cause mild symptoms upon primary infection and severe symptoms upon secondary infection (antibody-dependent enhancement). Estimating the number of unreported patients in hospitals and other facilities (which is difficult to obtain) is therefore important for predicting the magnitude of future epidemics. In collaboration with a research team on site that has been collecting dengue patient data for more than 30 years, and through long-term discussions with them, we have developed an algorithm to improve this situation. The algorithm estimates the distribution of unobserved patients from the distribution of actually observed patients 3.
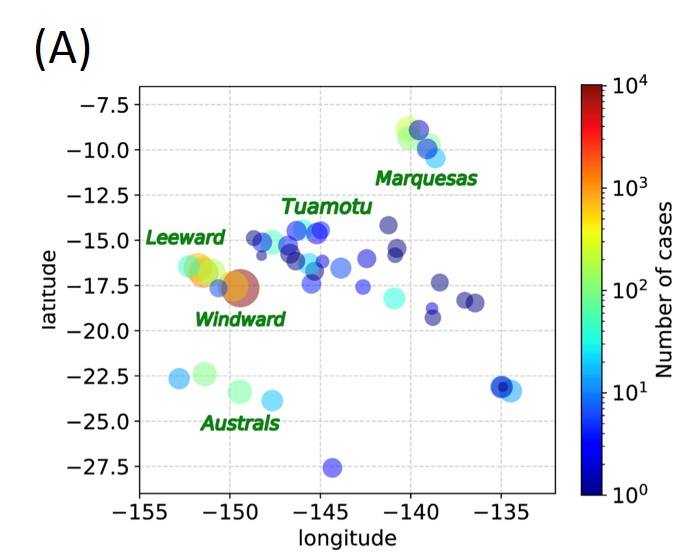
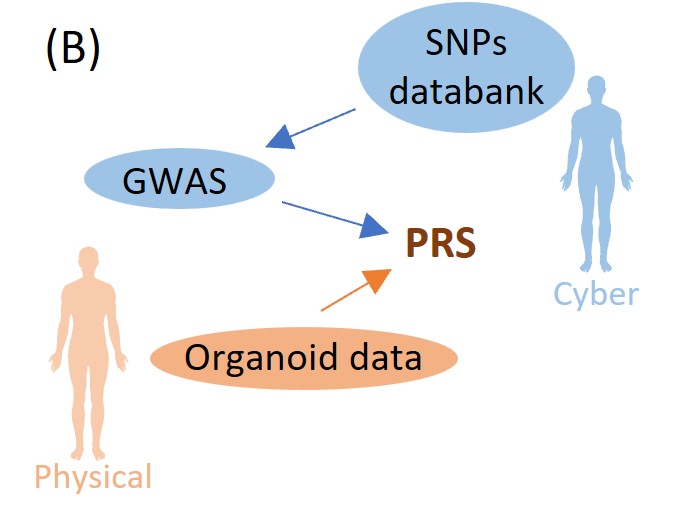
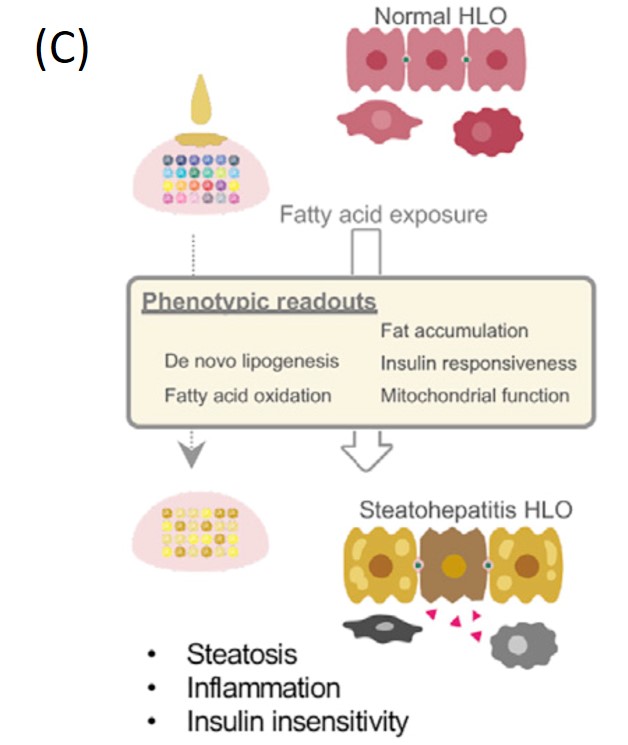
Fig. French Polynesia dengue patient distribution (A) 3, PRS calibrated by organoid experiments (B) and lipid accumulation in liver organoids (C) 5
A future plan
Generating a human organoid for each patient in a clinical setting is impractical. In human digital twins, it is therefore essential to introduce a patient “risk score” reflecting the results of human organoid experiments, (which can be calculated without actually performing the experiments on that patient). Genome-wide association studies (GWAS) identify effect sizes for single-nucleotide variants, from which a Polygenic Risk Score (PRS) can be calculated in order to evaluate each patient’s risk of disease: the basic idea is that the PRS is the sum of the effect sizes of the genetic variants that each patient possesses. PRS can be improved in a number of ways using target datasets in addition to the effect sizes and other summary statistics obtained from the GWAS, e.g., by reducing the number of variants considered (with clumping/pruning and thresholding) or by introducing correlation structures between different variants (Bayesian LDpred). In a similar manner, our goal is to fine-tune PRS by using the human organoid experimental data (Figure B). This is being studied in close collaboration with Prof. Takebe and his team, using the experimental data on lipid accumulation in liver organoids (Figure C) 5.
References