SHINOBU Ai
Specially Appointed Associate Professor (Full Time)/PI
PRIMe, The University of Osaka
- Related Website:
- ORCID research gate researchmap
Our researchers
PRIMe researchers from diverse fields of study, nationalities, and backgrounds come together and collaborate “under-one-roof” to conduct interdisciplinary and integrative research.

SHINOBU Ai
Specially Appointed Associate Professor (Full Time)/PI
PRIMe, The University of Osaka
Drug Discovery Methods Development and Applications Using Molecular Dynamics Simulations
Development of computational methods for modeling and simulating binding of drugs to their targets for drug discovery. Bridging the gap between the molecular and organoid scales by designing simulation methods for elucidating the molecular mechanism of protein-protein interactions.
Research Outline
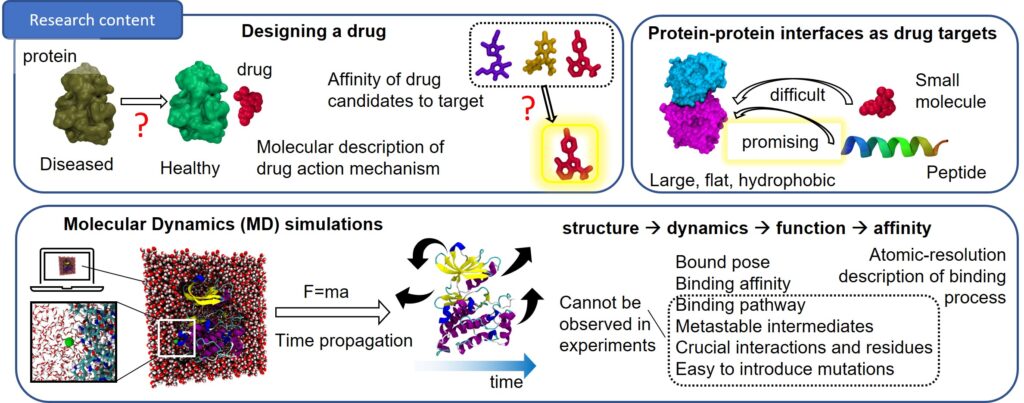
Research plan
Peptides as drug candidates
・Efficient sampling of binding/unbinding of protein-peptide (currently computationally difficult)
・Affinity-based screening of drug candidates using free-energy calculations and amino-acid mutations
MD simulation-based personalized medicine
・Identify the structural basis for variation in drug efficacy due to genomic variation among patients
・Design drugs that overcome drug-resistant mutations
Molecular-level description of protein-protein interactions (PPI)
・Identify the crucial elements of PPIs
・Develop multi-scale models for simulating dynamics of protein-protein complexes
Past research and preparation state
•Large-scale simulations of protein-inhibitor binding1
•Parameter tuning for obtaining a converged free-energy landscape and introduction of parameter setting protocol2
•Effect of inhibitor size and flexibility on the binding mechanism1
Determination of the correct binding pose of protein-protein complexes3,4
Coarse-grained model for sampling large conformational changes in proteins5,6
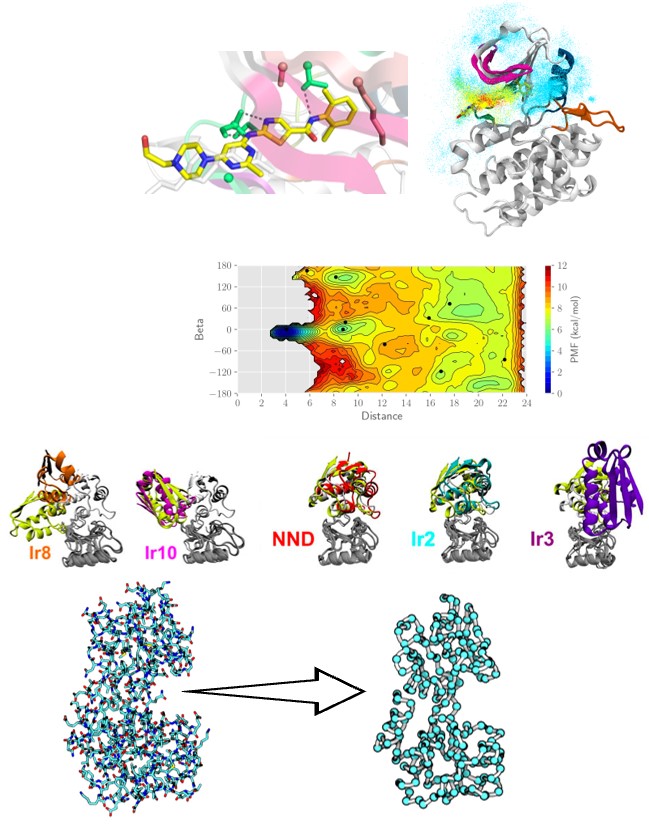

References