Imad ABUGESSAISA
Specially Appointed Professor (Full Time)/PI
PRIMe, The University of Osaka
- Related Website:
- PubMed Google Scholar researchmap KAKEN
Our researchers
PRIMe researchers from diverse fields of study, nationalities, and backgrounds come together and collaborate “under-one-roof” to conduct interdisciplinary and integrative research.

Imad ABUGESSAISA
Specially Appointed Professor (Full Time)/PI
PRIMe, The University of Osaka
Education background
Research Outline
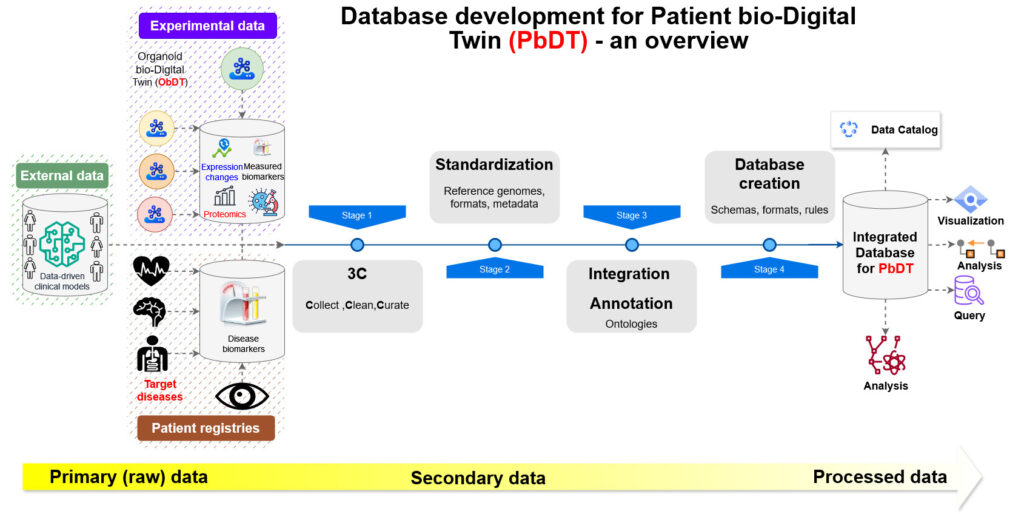
Figure【1】 An overview of database development and information flow for Patient bio-Digital Twin (PbDT). The database will contain raw (raw sequence reads) , secondary (mapped reads) and processed (summarized expression) data. Each of the four steps encompasses multiple steps and procedures.
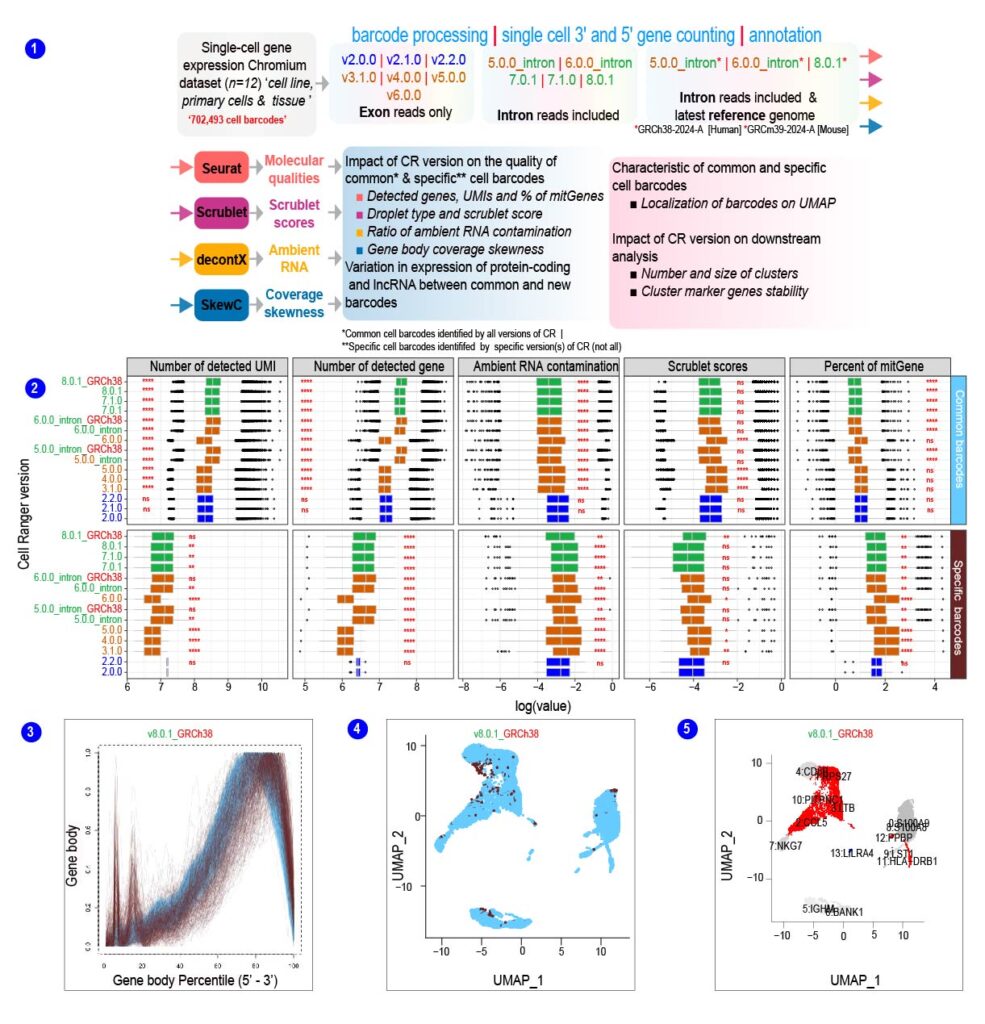
Figure【2】 Workflow for the QC and evaluation of chromium single cell gene expression data (Abugessaisa I, et al., Impacts of Cell Ranger versions on Chromium gene expression data bioRxiv 2024. [DOI]: https://doi.org/10.1101/2024.08.10.607413.
① Overview of the computation workflow and basic characteristics of cell barcodes generated by different versions of Cell Ranger pipeline. a. 8k PBMCs from a Healthy Donor single-cell gene expression chromium data. The dataset was generated with Single Cell 3′ v2 protocol. Cell barcodes were called 15 times by different versions of Cell Ranger (CR). The output from each version of CR was used as input for the scRNA-seq QC methods to compute molecular qualities, scrublet score, ambient RNA contamination, and gene body coverage. ② Significant differences between common and specific barcodes in molecular qualities determined by number of detected UMIs, genes, ratio of ambient RNA contamination, Scrublet scores, and percent of mitGenes. The upper panel shows common barcodes, and the lower panel shows specific barcodes. The mean comparison p-values added to the boxplot to indicate the statistical significance. ③ The gene body coverage was computed and visualized by SkewC. Each line represents a cell barcode. X-axis, gene body coverage percentile 5`-end to 3`-end, Y-axis was the gene body coverage. Gene body coverage plot colored by the type of the cell barcodes. The specific cell barcodes have peaks at the 5` ends of the gene region (skewed). ④ UMAP embedding of PBMC_8k cells colored by the source of the cell barcodes. Cell barcodes in light blue represent the cell barcodes called by all versions of Cell Ranger (common barcodes). Cell barcodes colored brown is the cell barcodes called by the specific Cell Ranger version (specific barcodes). ⑤ UMAP plot of the “common” cell barcodes. Each cluster in the UMAP is labeled with the cluster number and top marker gene. Unsupervised clusters in each UMAP are colored by the type of cluster markers. Red colored clusters indicate that the marker genes are changed from a previous CR version while the gray colored clusters indicate that the marker gene was conserved (not changed) from the previous version of CR.
References
Abugessaisa I, Hasegawa A, Walker S, Katayama S, Kere J, Kasukawa T. Impacts of Cell Ranger versions on Chromium gene expression data bioRxiv 2024. [DOI]: https://doi.org/10.1101/2024.08.10.607413
Abugessaisa I, Hasegawa A, Katayama S, Kere J, Kasukawa T. Computational approach to evaluate scRNA-seq data quality and gene body coverage with SkewC. STAR Protoc. (2023). [PMID]: 36853658.
Abugessaisa I, Hasegawa A, Noguchi S, Cardon M, Watanabe K, Takahashi M, Suzuki H, Katayama S, Kere J, Kasukawa T. SkewC: Identifying cells with skewed gene body coverage in single-cell RNA sequencing data. iScience. (2022). [PMID]: 35146392.
Abugessaisa I, Noguchi S, Hasegawa A, Kondo A, Kawaji H, Carninci P, Kasukawa T. refTSS: A Reference Data Set for Human and Mouse Transcription Start Sites. J Mol Biol. (2019). [PMID]: 31075273.
Woogeng IN, Kaczkowski B, Abugessaisa I, et al. al. Inducing human retinal pigment epithelium-like cells from somatic tissue. Stem Cell Reports. (2022). [PMID]: 35030321.
Abugessaisa I, Ramilowski JA, Lizio M, Severin J, Hasegawa A, Harshbarger J, Kondo A, Noguchi S, Yip CW, Ooi JLC, Tagami M, Hori F, Agrawal S, Hon CC, Cardon M, Ikeda S, Ono H, Bono H, Kato M, Hashimoto K, Bonetti A, Kato M, Kobayashi N, Shin J, de Hoon M, Hayashizaki Y, Carninci P, Kawaji H, Kasukawa T. FANTOM enters 20th year: expansion of transcriptomic atlases and functional annotation of non-coding RNAs. Nucleic Acids Res. (2021). [PMID]: 33211864
Abugessaisa I, Noguchi S, Hasegawa A, Kondo A, Kawaji H, Carninci P, Kasukawa T. refTSS: A Reference Data Set for Human and Mouse Transcription Start Sites. J Mol Biol. (2019). [PMID]: 31075273.
de Rie D, Abugessaisa I, Alam T, Arner E, Arner P, Ashoor H, Åström G, Babina M, Bertin N, Burroughs AM, Carlisle AJ, Daub CO, Detmar M, Deviatiiarov R, Fort A, Gebhard C, Goldowitz D, Guhl S, Ha TJ, Harshbarger J, Hasegawa A, Hashimoto K, Herlyn M, Heutink P, Hitchens KJ, Hon CC, Huang E, Ishizu Y, Kai C, Kasukawa T, Klinken P, Lassmann T, Lecellier CH, Lee W, Lizio M, Makeev V, Mathelier A, Medvedeva YA, Mejhert N, Mungall CJ, Noma S, Ohshima M, Okada-Hatakeyama M, Persson H, Rizzu P, Roudnicky F, Sætrom P, Sato H, Severin J, Shin JW, Swoboda RK, Tarui H, Toyoda H, Vitting-Seerup K, Winteringham L, Yamaguchi Y, Yasuzawa K, Yoneda M, Yumoto N, Zabierowski S, Zhang PG, Wells CA, Summers KM, Kawaji H, Sandelin A, Rehli M; FANTOM Consortium; Hayashizaki Y, Carninci P, Forrest ARR, de Hoon MJL. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat Biotechnol. (2017). [PMID]: 28829439.
Abugessaisa I*, Manabe RI, Kawashima T, Tagami M, Takahashi C, Okazaki Y, Bandinelli S, Kasukawa T, Ferrucci L. OVCH1 Antisense RNA 1 is differentially expressed between non-frail and frail old adults. Geroscience. (2024). [PMID]: 37817005
* Corresponding author
Gomez-Cabrero D, Walter S, Abugessaisa I, Miñambres-Herraiz R, Palomares LB, Butcher L, Erusalimsky JD, Garcia-Garcia FJ, Carnicero J, Hardman TC, Mischak H, Zürbig P, Hackl M, Grillari J, Fiorillo E, Cucca F, Cesari M, Carrie I, Colpo M, Bandinelli S, Feart C, Peres K, Dartigues JF, Helmer C, Viña J, Olaso G, García-Palmero I, Martínez JG, Jansen-Dürr P, Grune T, Weber D, Lippi G, Bonaguri C, Sinclair AJ, Tegner J, Rodriguez-Mañas L; FRAILOMIC initiative. A robust machine learning framework to identify signatures for frailty: a nested case-control study in four aging European cohorts. Geroscience. (2021). [PMID]: 33599920.
Education

Practical Guide to Life Science Databases, a textbook edited by Imad Abugessaisa and Takeya Kasukawa (2023). Singapore, Springer Nature 27–56.