HIMENO Yukiko
Specially Appointed Associate Professor (Full Time)
PRIMe, The University of Osaka
- Related Website:
- researchmap ORCID Loop
Our researchers
PRIMe researchers from diverse fields of study, nationalities, and backgrounds come together and collaborate “under-one-roof” to conduct interdisciplinary and integrative research.

HIMENO Yukiko
Specially Appointed Associate Professor (Full Time)
PRIMe, The University of Osaka
Reproducing Physiological Functions Mathematically in a Spatiotemporal Space
1. Simulating Physiology of Cardiomyocytes
The heart functions as a pump that circulates blood throughout the body by gathering contractile force of billions of electrically interconnected myocardial cells, forming a functional syncytium. Developing a mathematical model using ordinary differential equations based on electrophysiological properties revealed in experiment at the ionic and molecular level using isolated cardiomyocytes enabled us to simulate contractile force generation in myocardial cells dynamically [1]. By electrically coupling those multiple cells represented mathematically, it becomes possible to predict tissue-level behavior [2]. Integrating these cardiac models with multi-dimensional datasets using bioinformatics approaches facilitates the elucidation of mechanisms underlying cardiac pathologies such as heart failure and arrhythmias at the ionic, protein, and genetic levels—ultimately contributing to the development of preventive and therapeutic strategies.
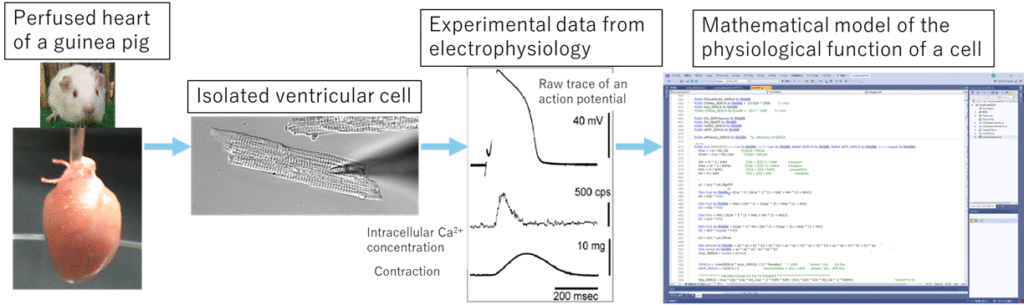
2. e-Heart Simulation Platform
Understanding the physiological behavior of cardiomyocytes requires a comprehensive understanding of their functional components, including ion channels and transporters involved in membrane ion transport, signal transduction pathways, and contractile proteins. This also requires fundamental knowledge in physical chemistry and thermodynamics and demands expertise. However, by applying informatics methodologies, it becomes feasible to construct simulators that predict cellular behavior by integrating modules that represent such components through mathematical formulations. The “e-Heart” educational simulator package [3,4], which defines parameters explicitly and simulates cellular activities, is now going to be adapted into a cloud-based platform to make it widely accessible to both students and researchers.
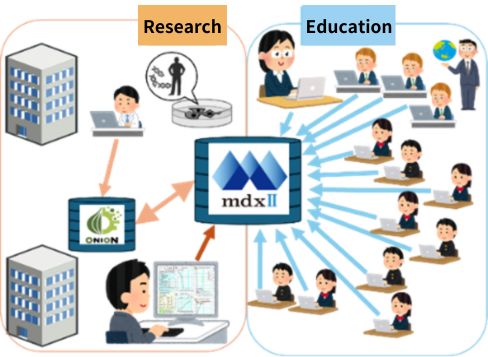
3. Envisioning the Future with Bio-Digital Twins
Development of bio-digital twins—virtual entities that replicate biological functions of a human within a mathematical spatiotemporal domain by constructing organ-specific simulators and tailoring them to individual characteristics, are underway. By combining these personalized organ simulators, virtual replicas that mirror biological individuality could be designed. To ensure these innovations lead to real-world health and wellbeing outcomes, active engagement from diverse stakeholders—including patients, citizens, and frontline professionals—is vital. Workshops will be held regularly to facilitate participatory development throughout the innovation process.

References